Virus
Virus classification is the process of naming viruses and placing them into a taxonomic system. Similar to the classification systems used for cellular organisms, virus classification is the subject of ongoing debate and proposals. This is mainly due to the pseudo-living nature of viruses, which is to say they are non-living particles with some chemical characteristics similar to those of life, or non-cellular life. As such, they do not fit neatly into the established biological classification system in place for cellular organisms.
Viruses are mainly classified by phenotypic characteristics, such as morphology, nucleic acid type, mode of replication, host organisms, and the type of disease they cause. The formal taxonomic classification of viruses is the responsibility of the International Committee on Taxonomy of Viruses (ICTV) system, although the Baltimore classification system can be used to place viruses into one of seven groups based on their manner of mRNA synthesis. Specific naming conventions and further classification guidelines are set out by the ICTV.
A catalogue of all the world’s known viruses has been proposed; some related preliminary efforts have been accomplished.[1]
Virus species definition
Edit
Species form the basis for any biological classification system. The ICTV had adopted the principle that a virus species is a polythetic class of viruses that constitutes a replicating lineage and occupies a particular ecological niche. In July 2013, the ICTV definition of species changed to state: “A species is a monophyletic group of viruses whose properties can be distinguished from those of other species by multiple criteria.”[2]
ICTV classification
Edit
The International Committee on Taxonomy of Viruses began to devise and implement rules for the naming and classification of viruses early in the 1970s, an effort that continues to the present. The ICTV is the only body charged by the International Union of Microbiological Societies with the task of developing, refining, and maintaining a universal virus taxonomy.[3]
The system shares many features with the classification system of cellular organisms, such as taxon structure. However, this system of nomenclature differs from other taxonomic codes on several points. A minor point is that names of orders and families are italicized,[4] unlike in the International Code of Nomenclature for algae, fungi, and plants and International Code of Zoological Nomenclature.
Viral classification starts at the level of realm and continues as follows, with the taxon suffixes given in italics[5]:
Realm (-viria)
Subrealm (-vira)
Kingdom (-viriae)
Subkingdom (-virites)
Phylum (-viricota)
Subphylum (-viricotina)
Class (-viricetes)
Subclass (-viricetidae)
Order (-virales)
Suborder (-virineae)
Family (-viridae)
Subfamily (-virinae)
Genus (-virus)
Subgenus (-virus)
Species
Species names often take the form of [Disease] virus, particularly for higher plants and animals. As of November 2018, only phylum, subphylum, class, order, suborder, family, subfamily, genus, and species are used.
The establishment of an order is based on the inference that the virus families it contains have most likely evolved from a common ancestor. The majority of virus families remain unplaced.
As of 2018, one realm, four incertae sedis orders, 46 incertae sedis families, and three incertae sedis genera are accepted:[6]
Realms: Riboviria
Incertae sedis orders: Caudovirales, Herpesvirales, Ligamenvirales, Ortervirales
Incertae sedis families: Adenoviridae, Alphasatellitidae, Ampullaviridae, Anelloviridae, Ascoviridae, Asfarviridae, Bacilladnaviridae, Baculoviridae, Bicaudaviridae, Bidnaviridae, Circoviridae, Clavaviridae, Corticoviridae, Fuselloviridae, Geminiviridae, Genomoviridae, Globuloviridae, Guttaviridae, Hepadnaviridae, Hytrosaviridae, Inoviridae, Iridoviridae, Lavidaviridae, Marseilleviridae, Microviridae, Mimiviridae, Nanoviridae, Nimaviridae, Nudiviridae, Ovaliviridae, Papillomaviridae, Parvoviridae, Phycodnaviridae, Plasmaviridae, Pleolipoviridae, Polydnaviridae, Polyomaviridae, Portogloboviridae, Poxviridae, Smacoviridae, Sphaerolipoviridae, Spiraviridae, Tectiviridae, Tolecusatellitidae, Tristromaviridae, Turriviridae
Incertae sedis genera: Dinodnavirus, Rhizidiovirus, Salterprovirus
Higher virus taxa span viruses with varying host ranges. The Ortervirales (Groups VI and VII), containing also retroviruses (infecting animals including humans e.g. HIV), retrotransposons (infecting invertebrate animals, plants and eukaryotic microorganisms) and caulimoviruses (infecting plants), are recent additions to the classification system orders.[7][8] Other variations occur between the orders: Nidovirales, for example, are isolated for their differentiation in expressing structural and nonstructural proteins separately.
Structure-based virus classification
Edit
It has been suggested that similarity in virion assembly and structure observed for certain viral groups infecting hosts from different domains of life (e.g., bacterial tectiviruses and eukaryotic adenoviruses or prokaryotic Caudovirales and eukaryotic herpesviruses) reflects an evolutionary relationship between these viruses.[9] Therefore, structural relationship between viruses has been suggested to be used as a basis for defining higher-level taxa – structure-based viral lineages – that could complement the existing ICTV classification scheme.[10]
Baltimore classification
Edit
Main articles: Baltimore classification and Virus Information Table
<img alt=”” src=”//upload.wikimedia.org/wikipedia/commons/thumb/0/07/Baltimore_Classification.png/300px-Baltimore_Classification.png” decoding=”async” width=”300″ height=”250″ class=”thumbimage” data-file-width=”831″ data-file-height=”692″>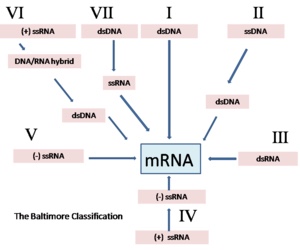
The Baltimore Classification of viruses is based on the method of viral mRNA synthesis
Baltimore classification (first defined in 1971) is a classification system that places viruses into one of seven groups depending on a combination of their nucleic acid (DNA or RNA), strandedness (single-stranded or double-stranded), sense, and method of replication. Named after David Baltimore, a Nobel Prize-winning biologist, these groups are designated by Roman numerals. Other classifications are determined by the disease caused by the virus or its morphology, neither of which are satisfactory due to different viruses either causing the same disease or looking very similar. In addition, viral structures are often difficult to determine under the microscope. Classifying viruses according to their genome means that those in a given category will all behave in a similar fashion, offering some indication of how to proceed with further research. Viruses can be placed in one of the seven following groups:[11]
• I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
• II: ssDNA viruses (+ strand or “sense”) DNA (e.g. Parvoviruses)
• III: dsRNA viruses (e.g. Reoviruses)
• IV: (+)ssRNA viruses (+ strand or sense) RNA (e.g. Picornaviruses, Togaviruses)
• V: (−)ssRNA viruses (− strand or antisense) RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
• VI: ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
• VII: dsDNA-RT viruses DNA with RNA intermediate in life-cycle (e.g. Hepadnaviruses)
<img alt=”” src=”//upload.wikimedia.org/wikipedia/commons/thumb/7/7c/The_Baltimore_Classification.gif/800px-The_Baltimore_Classification.gif” decoding=”async” width=”800″ height=”217″ class=”thumbimage” data-file-width=”2000″ data-file-height=”543″>
Visualization of the 7 groups of virus according to the Baltimore Classification
DNA viruses
Edit
Further information: DNA virus
• Group I: viruses possess double-stranded DNA. Viruses that cause chickenpox and herpes are found here.
• Group II: viruses possess single-stranded DNA.
Virus family
Examples (common names)
Virion
naked/enveloped
Capsid
symmetry
Nucleic acid type
Group
1. Adenoviridae
Adenovirus, infectious canine hepatitis virus
Naked
Icosahedral
ds
I
2. Papovaviridae
Papillomavirus, polyomaviridae, simian vacuolating virus
Naked
Icosahedral
ds circular
I
3. Parvoviridae
Parvovirus B19, canine parvovirus
Naked
Icosahedral
ss
II
4. Herpesviridae
Herpes simplex virus, varicella-zoster virus, cytomegalovirus, Epstein–Barr virus
Enveloped
Icosahedral
ds
I
5. Poxviridae
Smallpox virus, cow pox virus, sheep pox virus, orf virus, monkey pox virus, vaccinia virus
Complex coats
Complex
ds
I
7. Anelloviridae
Torque teno virus
Naked
Icosahedral
ss circular
II
7. Pleolipoviridae
HHPV1, HRPV1, HGPV1, His2V
Enveloped
ss/ds linear/circular
I/II
RNA viruses
Edit
Further information: RNA virus
• Group III: viruses possess double-stranded RNA genomes, e.g. rotavirus.
• Group IV: viruses possess positive-sense single-stranded RNA genomes. Many well known viruses are found in this group, including the picornaviruses (which is a family of viruses that includes well-known viruses like Hepatitis A virus, enteroviruses, rhinoviruses, poliovirus, and foot-and-mouth virus), SARS virus, hepatitis C virus, yellow fever virus, and rubella virus.
• Group V: viruses possess negative-sense single-stranded RNA genomes. The deadly Ebola and Marburg viruses are well known members of this group, along with influenza virus, measles, mumps and rabies.
Virus Family
Examples (common names)
Capsid
naked/enveloped
Capsid
Symmetry
Nucleic acid type
Group
1. Reoviridae
Reovirus, rotavirus
Naked
Icosahedral
ds
III
2. Picornaviridae
Enterovirus, rhinovirus, hepatovirus, cardiovirus, aphthovirus, poliovirus, parechovirus, erbovirus, kobuvirus, teschovirus, coxsackie
Naked
Icosahedral
ss
IV
3. Caliciviridae
Norwalk virus
Naked
Icosahedral
ss
IV
4. Togaviridae
Rubella virus, alphavirus
Enveloped
Icosahedral
ss
IV
5. Arenaviridae
Lymphocytic choriomeningitis virus
Enveloped
Complex
ss(-)
V
6. Flaviviridae
Dengue virus, hepatitis C virus, yellow fever virus, Zika virus
Enveloped
Icosahedral
ss
IV
7. Orthomyxoviridae
Influenzavirus A, influenzavirus B, influenzavirus C, isavirus, thogotovirus
Enveloped
Helical
ss(-)
V
8. Paramyxoviridae
Measles virus, mumps virus, respiratory syncytial virus, Rinderpest virus, canine distemper virus
Enveloped
Helical
ss(-)
V
9. Bunyaviridae
California encephalitis virus, hantavirus
Enveloped
Helical
ss(-)
V
10. Rhabdoviridae
Rabies virus
Enveloped
Helical
ss(-)
V
11. Filoviridae
Ebola virus, Marburg virus
Enveloped
Helical
ss(-)
V
12. Coronaviridae
Corona virus
Enveloped
Helical
ss
IV
13. Astroviridae
Astrovirus
Naked
Icosahedral
ss
IV
14. Bornaviridae
Borna disease virus
Enveloped
Helical
ss(-)
V
15. Arteriviridae
Arterivirus, equine arteritis virus
Enveloped
Icosahedral
ss
IV
16. Hepeviridae
Hepatitis E virus
Naked
Icosahedral
ss
IV
Reverse transcribing viruses
Edit
• Group VI: viruses possess single-stranded RNA viruses that replicate through a DNA intermediate. The retroviruses are included in this group, of which HIV is a member.
• Group VII: viruses possess double-stranded DNA genomes and replicate using reverse transcriptase. The hepatitis B virus can be found in this group.
Virus Family
Examples (common names)
Capsid
naked/enveloped
Capsid
Symmetry
Nucleic acid type
Group
1. Retroviridae
HIV
Enveloped
dimer RNA
VI
2. Caulimoviridae
Caulimovirus, Cacao swollen-shoot virus (CSSV)
Naked
VII
3. Hepadnaviridae
Hepatitis B virus
Enveloped
Icosahedral
circular, partially ds
VII
Holmes classification
Edit
Holmes (1948) used Carl Linnaeus’s system of binomial nomenclature to classify viruses into 3 groups under one order, Virales. They are placed as follows:
• Group I: Phaginae (attacks bacteria)
• Group II: Phytophaginae (attacks plants)
• Group III: Zoophaginae (attacks animals)
LHT System of Virus Classification
Edit
The LHT System of Virus Classification is based on chemical and physical characters like nucleic acid (DNA or RNA), symmetry (helical or icosahedral or complex), presence of envelope, diameter of capsid, number of capsomers.[12] This classification was approved by the Provisional Committee on Nomenclature of Virus (PNVC) of the International Association of Microbiological Societies (1962).[citation needed] It is as follows:
• Phylum Vira (divided into 2 subphyla)
• Subphylum Deoxyvira (DNA viruses)
• Class Deoxybinala (dual symmetry)
• Order Urovirales
• Family Phagoviridae
• Class Deoxyhelica (helical symmetry)
• Order Chitovirales
• Family Poxviridae
• Class Deoxycubica (cubical symmetry)
• Order Peplovirales
• Family Herpesviridae (162 capsomeres)
• Order Haplovirales (no envelope)
• Family Iridoviridae (812 capsomeres)
• Family Adenoviridae (252 capsomeres)
• Family Papiloviridae (72 capsomeres)
• Family Paroviridae (32 capsomeres)
• Family Microviridae (12 capsomeres)
• Subphylum Ribovira (RNA viruses)
• Class Ribocubica
• Order Togovirales
• Family Arboviridae
• Order Tymovirales
• Family Napoviridae
• Family Reoviridae
• Class Ribohelica
• Order Sagovirales
• Family Stomataviridae
• Family Paramyxoviridae
• Family Myxoviridae
• Order Rhabdovirales
• Suborder Flexiviridales
• Family Mesoviridae
• Family Peptoviridae
• Suborder Rigidovirales
• Family Pachyviridae
• Family Protoviridae
• Family Polichoviridae
Subviral agents
Edit
The following agents are smaller than viruses and have only some of their properties.
Viroids
Edit
Main article: Viroid
• Family Avsunviroidae[13]
◦ Genus Avsunviroid; type species: Avocado sunblotch viroid
◦ Genus Pelamoviroid; type species: Peach latent mosaic viroid
◦ Genus Elaviroid; type species: Eggplant latent viroid
• Family Pospiviroidae[14]
◦ Genus Pospiviroid; type species: Potato spindle tuber viroid
◦ Genus Hostuviroid; type species: Hop stunt viroid
◦ Genus Cocadviroid; type species: Coconut cadang-cadang viroid
◦ Genus Apscaviroid; type species: Apple scar skin viroid
◦ Genus Coleviroid; type species: Coleus blumei viroid 1
Satellites
Edit
Main article: Satellite (biology)
Satellites depend on co-infection of a host cell with a helper virus for productive multiplication. Their nucleic acids have substantially distinct nucleotide sequences from either their helper virus or host. When a satellite subviral agent encodes the coat protein in which it is encapsulated, it is then called a satellite virus.
• Satellite viruses[15]
◦ Single-stranded RNA satellite viruses
▪ Subgroup 1: Chronic bee-paralysis satellite virus
▪ Subgroup 2: Tobacco necrosis satellite virus
◦ Double-stranded DNA satellite viruses (virophages)
• Satellite nucleic acids
◦ Single-stranded satellite DNAs
◦ Double-stranded satellite RNAs
◦ Single-stranded satellite RNAs
▪ Subgroup 1: Large satellite RNAs
▪ Subgroup 2: Small linear satellite RNAs
▪ Subgroup 3: Circular satellite RNAs (virusoids)
Prions
Edit
Prions, named for their description as “proteinaceous and infectious particles”, lack any detectable (as of 2002) nucleic acids or virus-like particles. They resist inactivation procedures that normally affect nucleic acids.[16]
• Mammalian prions:
◦ Agents of spongiform encephalopathies
• Fungal prions:
◦ PSI+ prion of Saccharomyces cerevisiae
◦ URE3 prion of Saccharomyces cerevisiae
◦ RNQ/PIN+ prion of Saccharomyces cerevisiae
◦ Het-s prion of Podospora anserina
Defective interfering particles
Edit
Main article: Defective interfering particle
• Defective interfering RNA
• Defective interfering D











